Chapter 6, Basic Concepts of Enzyme Action
Chapter 7, Kinetics and Regulation
Chapter 8, Mechanisms and Inhibitors
Chapter 9, Hemoglobin, An Allosteric Protein
Biochemistry: A Short Course, 2nd ED.
| 471396701 | Enzymes are proteins that speed up the rate of chemical reactions. | Substrate -> Product *NOTE: Amount of product is the same, but an enzyme makes it faster. | |
| 471396702 | ΔG | The change in free energy for conversion of a substrate to product. | |
| 471396703 | X≠ | The transition state a reaction must go through. Substrate -> X≠ -> Product | |
| 471396704 | Equation between transition state and substrate. | ΔGx≠ - ΔGsubstrate = ΔG≠ | |
| 471396705 | What does the enzyme do, in relation to the transition state? | Enzymes stabilize the transition state, decreasing the energy barrier for product formation. | |
| 471396706 | The first step in catalysis. | Binding of the substrate to the enzyme in the active site. (Alpha helix Beta sheet turns) | |
| 471396707 | Active site structure. | In primary structure, a 3-D cleft formed by AA. | |
| 471396708 | What is the specificity of substrate binding to the active site dependent on? | The AA | |
| 471396709 | The substrate must have a matching shape to fit the active site. | ... |  |
| 471396710 | Two kinds of substrate/enzyme binding. | Lock and Key. Induced Fit. |  |
| 471396711 | The six major classes of enzymes. Their function? | 1. Oxidoreductase: catalyze redox reactions. 2. Transferase: move func.groups between molecules. 3. Hydrolyases: cleave bonds with the add. of H2O. 4. Lyases: remove atoms to form double bonds or add atoms to double bonds. 5. Isomerases: move func.groups within molecule. 6. Ligases: join two molecules at the expense of ATP. | |
| 471396712 | What are enzymes named for? | Named for their substrates and for the reaction they catalyze with suffix "ase" (IE. Peptide hydrolase) | |
| 471396713 | Cofactors | Small organic molecules and/or metals. | |
| 471396714 | Holoenzyme | Enzyme with its cofactor. | |
| 471396715 | Apoenzyme | Enzyme without its cofactor. | |
| 471396716 | Primary function of enzymes. | To accelerate the rates of reactions, so they are compatible with the needs of the organism. | |
| 471396717 | Kinetics | The study of the rates (velocities) of reactions. | |
| 471396718 | What do rates (velocities) depend on? | The concentration of substrate(s). | |
| 471396719 | Velocity (V) equation. | V= -Δsubstrate/Δtime =Δproduct/Δtime (decrease in substrate/time =increase in product/time) | |
| 471396720 | V=k[sub]^1 | k: rate constant, dependent on temperature V=1 M/s, [sub]=1M V=2 M/s, [sub]=2M double [sub], velocity doubles First order reaction | |
| 471396721 | Example Problems for V=k[sub]^1 | ex. |  |
| 471396722 | Investigation of enzyme kinetics. | Measure the initial velocity as a function of substrate concentration with a fixed amount of enzyme. (Compare kinetics between enzymes, substrates using the initial velocity, V0) | |
| 471501433 | Michaelis-Menten equation | Describes the variation of enzyme activity as a function of substrate concentration. V0= Vmax * [S]/[S]+Km |  |
| 471501434 | Km | Affinity for substrate. |  |
| 471501435 | Vmax | More efficient catalyst. | |
| 471501436 | Example problems for Michaelis-Menten. | On powerpoint (Lecture 09-18), slide 16. |  |
| 471501437 | Lineweaver-Burke equation | M-M equation can be manipulated into one that yields a straight-line plot. This is Double-reciprocal equation. kcat= Vmax/[enzyme] | |
| 471501438 | kcat | mx # of molecules an enzyme can "turn over" to product per second. | |
| 471501439 | kcat/Km | Measure of catalytic efficiency because it takes into account both the rate of catalysis (kcat) and nature of the enzyme substrate interaction (Km). Can be used to compare kinetic data for different substrates. | |
| 471501440 | High Km | Implies weak binding. | |
| 471501441 | Low Km | Implies strong binding. | |
| 472265959 | Acetaldehyde is more toxic than ethanol. | Sensitivity to ethanol is caused by increased amount of acetaldehyde (a mutation in ALDH2). | |
| 472265960 | How many forms of aldehyde dehydrogenase are there in the body? | Two. | |
| 472265961 | The two forms of aldehyde dehydrogenase in the body: | 1. Mitochondrial, low Km 2. Cytoplasmic, high Km | |
| 472265962 | Persons with sensitivity to ethanol have defect in what enzyme? | Mitochondrial enzyme, so only cytoplasmic enzyme works. | |
| 472265963 | Why is cytoplasmic enzyme efficient only at high [aldehyde]? | Because it is high Km. | |
| 472265964 | Tyrosinase | Involved in synthesizing pigment and has a low tolerance for heat. IE. The extremities of a Siamese cat are cool enough for tyrosinase to be active and produce pigment. | |
| 472265965 | Enzymes have an optimal pH. | ... |  |
| 472265966 | Enzyme reversible inhibition types | *Can bind and dissociate from the enzyme. 1. Competitive inhibition 2. Uncompetitive inhibition 3. Noncompetitive inhibition | |
| 472265967 | Enzyme inhibition can be reversible or irreversible. | ... | |
| 472265968 | Competitive Inhibition | Inhibitor resembles the substrate and binds to the active site. This reduces catalytic rate by reducing [enzyme-substrate]. |  |
| 472265969 | Inhibition can be relieved by increasing substrate. | ... |  |
| 472265970 | In competitive inhibition, Vmax does not change; Km increases. | ... |  |
| 472265971 | Uncompetitive Inhibition | Inhibitor binds only to the enzyme-substrate complex, and cannot be overcome by addition of substrate. |  |
| 472265972 | In uncompetitive inhibition, both Vmax and Km decrease. | ... |  |
| 472265973 | Noncompetitive Inhibition | Inhibitor and substrate bind simultaneously to the enzyme at different sites. This decreases active enzyme molecules, and cannot be overcome by increasing substrate. | |
| 472311159 | In noncompetitive inhibition, Vmax decreases and Km is unchanged. | ... |  |
| 472311160 | Irreversible Inhibitors | Bind very tightly to enzymes and do not readily dissociate. 1. Group specific reagents- react with specific R groups of AA. 2. Affinity labels- more specific for active site than group specific reagents. 3. Suicide inhibitors- chemically modified substrates. | |
| 472311161 | Irreversible inhibitors are good for cloning, bad for curing bacterial infections in people. | ... | |
| 472311162 | Like in noncompetitive inhibition, irreversible inhibitors have Vmax decrease and Km unchanged. | ... | |
| 472311163 | Allosteric Enzymes | Change their conformation upon binding of an effector, which results in an apparent change in binding affinity at a different ligand binding site. |  |
| 472311164 | Lack of allosteric inhibition leads to accumulation of uric acid. | ... | |
| 472311165 | Allosterically regulated enzymes do not conform to Michaelis-Menten Kinetics. | The reaction velocity of allosteric enzymes displays a sigmoidal relationship to substrate concentration. | 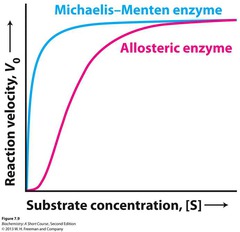 |
| 472311166 | Allosteric enzymes depend on alterations in 4ternary structure. | Allosteric enzymes can display 4ternary structure with multiple active sites and regulatory sites. | |
| 472311167 | Cooperativity | The binding of substrate to one cative site causes a conformation change. This change induces a change at a second subunit. | |
| 472311168 | Cooperativity display | It is displayed by enzymes with multiple binding sites where affinity of the binding sites for a ligand is increased, positive cooperativity, or decreased, negative cooperativity, upon the binding of a ligand to a binding site. | |
| 472311169 | Allostery plays a crucial role in metabolism (and many other fundamental biological processes). | ... | |
| 472311170 | Phosphofructokinase (in the Glycolytic cycle) is an allosteric enzyme. | PFK-1 activity increases with increasing ATP, but a point is reached where ATP inhibits activity. | |
| 472311171 | How can ATP be a substrate and inhibitor? | ATP can be a substrate and an allosteric regulator. | |
| 472311172 | How can glycolysis be regulated by ATP? | If [ATP] is high, PFK would be inhibited, glycolysis inhibited. | |
| 472311173 | Hemoglobin is a RBC protein that carries oxygen from the lungs to the tissues. | It is an allosteric protein that displays cooperativity in oxygen binding and release. | |
| 472311174 | Myoglobin binds oxygen in muscle cells. | This binding is not cooperative. Oxygen binding is measured as a function of the partial pressure of oxygen (pO2). | |
| 472311175 | Hemoglobin displays 4ternary structure with 4 active sites. | ... | |
| 472311176 | What does the heme group in hemoglobin consist of? | An organic compound (protoporphyrin) and a central iron ion in the ferrous (Fe2+) form. | |
| 472311177 | The iron in the heme group lies in the middle of the protoporphyrin bound to 4 nitrogens. | ... |  |
| 472311178 | Iron's two additional bonds | Fifth and Sixth coordination sites. |  |
| 472311179 | What happens upon oxygen binding? | The iron moves into the plane of the protoporphyrin ring. | |
| 472311180 | The movement of Fe leads to increased affinity for O2 binding at 2nd site. | ... | |
| 472311181 | What controls the affinity of O2 by hemoglobin? | Allosteric regulator. 2,3-bisphophoglycerate binds to Hb reducing its O2 affinity promoting O2 release. | |
| 472311182 | Cardon dioxide and H+, produced by actively respiring tissues, enhance oxygen release by hemoglobin. | ... |  |
| 472311183 | Salt Bridge | Positive and negative charges interacting. H+ and CO2 stabilize deoxyHb through salt bridges. | |
| 472311184 | Why can you die from CO poisoning? | CO has a 210-fold higher affinity for hemoglobin compared with oxygen. CO binding also prevents hemoglobin from acquiring CO2 from tissues for removal and stabilizes oxygen molecules bound to the same hemoglobin protein, preventing their release to tissues. |  |

