IB Bio (HL)
| 8695811221 | Metabolism | Sum total of all reactions that occur within an organism in order to maintain life | 0 | |
| 8695823212 | Metabolic pathways | Series of reactions in cells controlling chemical changes through specific enzymes | 1 | |
| 8695843557 | Examples of metabolic chains | Glycolysis (in cell respiration), coagulation cascade (in blood clotting) | 2 | |
| 8695845443 | Examples of metabolic cycles | Krebs cycle (in cell respiration), Calvin cycle (in photosynthesis) | 3 | |
| 8695851156 | Activation energy (EA) | Certain amount of energy required for chemical reactions to proceed | 4 | |
| 8695856041 | Enzyme purpose | Lowers activation energy | 5 | |
| 8695860149 | Process of enzyme catalysis | 1) Enzyme binds to substrate, stressing and destabilizing its the bonds in the substrate 2) Resulting in reduced overall energy level of substrates transitionary state | 6 | |
| 8695879267 | Types of enzyme reactions | Exergonic and endergonic | 7 | |
| 8695881850 | Exergonic reaction | Reactants contain more energy than products, free energy is released into system - Usually catabolic, as energy is released from broken bonds within a molecule | 8 | |
| 8695882806 | Endergonic reaction | Reactants contain less energy than products, free energy is lost to system - Usually anabolic, as energy is required to synthesise bonds between molecules | 9 | |
| 8695914577 | Enzyme inhibitor | Molecule that disrupts the normal reaction pathway between an enzyme and a substrate - Competitive / non-competitive, depending on their mechanism of action | 10 | |
| 8695934744 | Types of Enzyme Inhibition | - Normal enzyme reaction - Competitive Inhibition - Noncompetitive Inhibition | 11 | |
| 8695938420 | 2 Examples of Enzyme Inhibition | - Relenza (Competitive Inhibitor) - Cyanide (Noncompetitive Inhibitor) | 12 | |
| 8695941601 | 5 steps of Normal Enzyme Reaction | 1) Substrate binds to enzyme (via the active site) to form an enzyme-substrate complex 2) Shape and properties of the substrate and active site are complementary, resulting in enzyme-substrate specificity 3) Active site undergoes conformational change to optimally interact with the substrate (induced fit) 4) This change destabilised substrate chemical bonds substrate, lowering activation energy 5) Substrate is converted into product at an accelerated rate | 13 | |
| 8695956025 | Normal enzyme reaction diagram |  | 14 | |
| 8695959363 | Competitive Inhibition | Molecule, other than the substrate, binding to the enzyme's active site | 15 | |
| 8695960786 | 3 steps of Competitive Inhibition | 1) Molecule (inhibitor) is structurally and chemically similar to substrate (--> able to bind to active site) 2) Competitive inhibitor blocks active site and thus prevents substrate binding 3) As inhibitor competes with substrate, its effects can be reduced by increasing substrate concentration | 16 | |
| 8695973281 | Noncompetitive Inhibition | Molecule binding to a site other than the active site (allosteric site) | 17 | |
| 8695980457 | Competitive Inhibition diagram |  | 18 | |
| 8695974304 | 3 steps of Noncompetitive Inhibition | 1) Binding of inhibitor to allosteric site causes conformational change to enzyme's active site 2) Resulting in active site and substrate no longer sharing specificity, (substrate cannot bind) 3) As inhibitor isn't competing with substrate, increasing substrate levels cannot mitigate the inhibitor's effect | 19 | |
| 8695982642 | Noncompetitive Inhibition diagram |  | 20 | |
| 8695991512 | An example of a use for a competitive inhibitor | Treatment of influenza via the neuraminidase inhibitor, Relenza | 21 | |
| 8695994697 | Example of a use for a non-competitive inhibitor | Use of cyanide as a poison (prevents aerobic respiration) | 22 | |
| 8696009481 | Relenza (Competitive Inhibitor) | Synthetic drug designed by Australian scientists to treat individuals infected with the influenza virus | 23 | |
| 8696014156 | 3 steps of Relenza's inhibition | 1) Release of Virions from infected cells when viral enzyme neuraminidase cleaves docking protein (haemagglutinin) 2)Relenza competitively binds to neuraminidase active site, preventing cleavage of docking protein 3) --> virions aren't released from infected cells, preventing spread of influenza virus | 24 | |
| 8696025300 | Cyanide (Noncompetitive Inhibitor) | Poison which prevents ATP production via aerobic respiration, leading to eventual death | 25 | |
| 8696064487 | 3 steps of Cyanide inhibition | 1) Binds to allosteric site on cytochrome oxidase 2) By changing the shape of the active site, cytochrome oxidase can no longer pass electrons to the final acceptor (oxygen) 3) --> electron transport chain cannot continue to function and ATP is not produced via aerobic respiration | 26 | |
| 8696075097 | Cytochrome oxidase | A carrier molecule that forms part of the electron transport chain | 27 | |
| 8696086095 | End-product inhibition (or feedback inhibition) | Form of negative feedback by which metabolic pathways can be controlled, where the final product in a series of reactions inhibits an enzyme from an earlier step in the sequence | 28 | |
| 8710827965 | Two steps of end-product inhibition (or feedback inhibition) | 1) Product binds to allosteric site and temporarily inactivates enzyme (via non-competitive inhibition) 2) As enzyme can't function, reaction sequence is halted and rate of product formation decreases | 29 | |
| 8710839795 | End-product inhibition (or feedback inhibition) function | Ensure levels of an essential product are always tightly regulated | 30 | |
| 8710849908 | How End-product inhibitions (or feedback inhibition) regulate levels of essential products | - Product levels build up, product inhibits reaction pathway --> decreasing rate of product formation - Product levels drop, reaction pathway proceeds unhindered --> rate of product formation increases | 31 | |
| 8710863702 | End-Product Inhibition diagram |  | 32 | |
| 8710868716 | Isoleucine | 1 Essential amino acid, (is not synthesised by human body --> and must be ingested) | 33 | |
| 8710876067 | Food sources rich in isoleucine | Eggs, seaweed, fish, cheese, chicken and lamb | 34 | |
| 8710889061 | 3 steps of Isoleucine production in plants and bacteria | 1) Threonine converted into intermediate compound by an enzyme (threonine deaminase) 2) Isoleucine can bind to allosteric site on this enzyme, functioning as a non-competitive inhibitor 3) As excess production of isoleucine inhibits further synthesis, it functions as an example of end-product inhibition | 35 | |
| 8726740102 | Distinguishing different types of inhibition from graphs at specified substrate concentrations |  | 36 | |
| 8726751772 | Malaria | Disease caused by parasitic protozoans of the genus Plasmodium | 37 | |
| 8726769822 | How new anti-malarial drugs and medications can be produced | By targeting enzymes for inhibition, which coordinate the development of the parasite in both human and mosquito. | 38 | |
| 8726819084 | How scientists have identified potential targets for inhibition | 1) Sequenced genome of Plasmodium and used it to determine the parasite's proteome 2) Identified enzymes involved in parasitic metabolism from the proteome 3) screened enzymes against bioinformatic databases of chemicals to identify potential enzyme inhibitors 4) Chemically modified promising compounds to improve binding affinity and lower its toxicity. (Or by rational drug design) | 39 | |
| 8726874721 | Rational drug design | Using computer modelling techniques to invent compound that will function as an inhibitor: - Using combinatorial chemistry, a compound is synthesised that is complementary to the active site of the target enzyme | 40 | |
| 8726955481 | Adenosine triphosphate (ATP) | High energy molecule that functions as an immediate power source for cells |  | 41 |
| 8726972285 | How ATP stores energy | The three covalently bonded phosphate groups per molecule store potential energy in their bonds | 42 | |
| 8727044409 | How ATP releases energy | When ATP is hydrolyzed (--> ADP + Pi) energy stored in phosphate bond is released for cell use | 43 | |
| 8727052206 | ATP energy release diagram |  | 44 | |
| 8727059566 | Two key functions of ATP within cell | - Energy currency of cell by releasing energy when hydrolysed to ADP (powers cell metabolism) - May transfer released phosphate group to other organic molecules, rendering them less stable and more reactive | 45 | |
| 8727095892 | Two sources of energy from which ATP is synthesized from ADP | Solar energy and oxidative processes | 46 | |
| 8727103738 | How solar energy provides energy for ATP synthesis from ADP | Converts light energy into chemical energy that is stored as ATP | 47 | |
| 8727109320 | How oxidative processes provide energy for ATP synthesis from ADP | Cell respiration breaks down organic molecules to release chemical energy that is stored as ATP | 48 | |
| 8727157382 | Cell respiration | Controlled release of energy from organic compounds to produce ATP | 49 | |
| 8727164030 | Anaerobic respiration | Incomplete breakdown of organic molecules for a small yield of ATP (no oxygen required) | 50 | |
| 8727168360 | Aerobic respiration | Complete breakdown of organic molecules for a larger yield of ATP (oxygen is required) | 51 | |
| 8727181697 | Processes via which the breakdown of organic molecules occur | - Direct combustion of sugar - Stepwise oxidation of sugar (cell respiration) | 52 | |
| 8727231185 | Diagram of Energy Conversions in Sugar Breakdown (Direct Combustion vs Cell Respiration) |  | 53 | |
| 8727240290 | Redox Reactions | Reduction of one chemical species and the oxidation of another (redox = reduction / oxidation) | 54 | |
| 8727250132 | Reduction | gain of electrons / hydrogen or the loss of oxygen | 55 | |
| 8727250133 | Oxidation | loss of electrons / hydrogen or the gain of oxygen | 56 | |
| 8727318938 | What carries the energy released by oxidation reactions to the cristae of the mitochondria? | NAD and FAD | 57 | |
| 8727323527 | Energy Transfer via Hydrogen Carriers diagram |  | 58 | |
| 8727336997 | Why is cell respiration a oxidation reaction | Cell respiration breaks down organic molecules and transfers hydrogen atoms and electrons to carrier molecules (NAD and FAD) | 59 | |
| 8727364858 | Carrier molecules of cellular respiration nicknames | Hydrogen carriers or electron carriers | 60 | |
| 8727376341 | Most common hydrogen carrier | NAD+ | 61 | |
| 8727386426 | NAD + is reduced to | NADH | 62 | |
| 8727392322 | FAD is reduced to | FADH2 | 63 | |
| 8727381647 | A less common hydrogen carrier | FAD | 64 | |
| 8727396098 | Cristae | Site of electron transport chain, which uses energy transferred by the carriers to synthesize ATP | 65 | |
| 8727417515 | What respiration can generate ATP from hydrogen carriers | Aerobic, as electron transport requires oxygen to function | 66 | |
| 8727461544 | Why does aerobic respiration unlock more energy stored in the organic molecules and produce more ATP? | Because Oxygen is required for electron transport | 67 | |
| 8726980264 | Phosphorylation | Process making molecules less stable, which for example breaks down the ATP bonds, which are readily reactive. | 68 | |
| 8726696482 | Isoleucine production in plants and bacteria diagram |  | 69 | |
| 8728364779 | Organic compounds that can be used in cell respiration | Lipids, Proteins but mainly Carbohydrates | 70 | |
| 8728371136 | Reason why lipids are not preferentially used | They are harder to transport and digest (Although they yield more energy) | 71 | |
| 8728376987 | Reason why proteins are not preferentially used | They release potentially toxic nitrogenous compounds when broken down | 72 | |
| 8728434976 | Glycolysis | Breakdown of a hexose sugar (6C) into 3 pyruvate molecules (3C) | 73 | |
| 8728443441 | 4 Key events of Glycolysis | 1) Phosphorylation 2) Lysis 3) Oxidation 4) ATP formation | 74 | |
| 8728484902 | Phosphorylation in Glycolysis | Hexose sugar is phosphorylated by two molecules of ATP (to form a hexose bisphosphate) (makes molecule less stable and more reactive, and prevents diffusion out of the cell) | 75 | |
| 8728498160 | Lysis in Glycolysis | Hexose bisphosphate (6C sugar) is split into two triose phosphates (3C sugars) | 76 | |
| 8728503876 | Oxidation in Glycolysis | Removal of hydrogen atoms from each of the 3C sugards (via oxidation) to reduce NAD+ to NADH (In total 2 molecules of NADH are produced) | 77 | |
| 8728503877 | ATP formation in Glycolysis | Some energy released from the sugar intermediates is used to directly make ATP (In total 4 molecules of ATP are generated) | 78 | |
| 8728549337 | Overview of Glycolysis |  | 79 | |
| 8728580483 | Glycolysis occurs in | The cytosol | 80 | |
| 8728585709 | Is Glycolysis aerobic or anaerobic | Anaerobic, as no oxygen is required | 81 | |
| 8728616518 | Fermentation | - If no oxygen is present, the pyruvate remains in the cytosol and is converted into lactic acid (animals) or ethanol and CO2 (Plants and yeast) - A reduction reaction that oxidises NADH allows small amounts of ATP to be produced w.out oxyten | 82 | |
| 8728667323 | Anaerobic Respiration (Fermentation) |  | 83 | |
| 8728941509 | First stage of aerobic cell respiration | Link reaction | 84 | |
| 8728944965 | Link reaction | Transports pyruvate into the mitochondria | 85 | |
| 8728966028 | 4 stages of the Link reaction | 1) Pyruvate is transported from cytosol into mitochondrial matrix by carrier proteins on mitochondrial membrane 2) Pyruvate loses a carbon atom (decarboxylation), which forms a carbon dioxide molecule 3) The 2C compound then forms acetyl group when it loses hydrogen atoms via oxidation (NAD+ is reduced to NADH + H+) 4) Acetyl compound combines with coenzyme A to form acetyl coenzyme A (acetyl CoA) | 86 | |
| 8728995091 | How often per molecule of glucose does the link reaction occur? | Twice, due to glucose being split into two pyruvate molecules | 87 | |
| 8729005880 | Link reaction product per reaction | Acetyl CoA (2x) + H+ (2x) + CO2 (2x) | 88 | |
| 8729014257 | Diagram of the Link Reaction |  | 89 | |
| 8729026144 | Second stage of aerobic respiration | The Krebs cycle | 90 | |
| 8729029584 | The Krebs cycle occurs in | The matrix of the mitochondria | 91 | |
| 8729084768 | The Krebs | Transfer of acetyl group to a 4C compound to make a 6C compound - Coenzyme A is released and can return to the link reaction to form another molecule of acetyl CoA | 92 | |
| 8729145764 | 3 steps of 6C compound breakdown to form the original 4C compound | 1) Two C atoms are released via decarboxylation to form two molecules of carbon dioxide (CO2) 2) Multiple oxidation reactions result in the reduction of hydrogen carriers (3 × NADH + H+ ; 1 × FADH2) 3) One molecule of ATP is produced directly via substrate level phosphorylation | 93 | |
| 8729166773 | How many times does the Krebs cycle occur per molecule? | Twice, as the link reaction produces two molecules of acetyl CoA (1 per pyruvate) | 94 | |
| 8729174328 | Krebs cycle product per cycle | 4 × CO2 ; 2 × ATP ; 6 × NADH + H+ ; 2 × FADH2 | 95 | |
| 8729182391 | The Krebs Cycle diagram |  | 96 | |
| 8729208365 | Final stage of aerobic respiration | Electron transport chain | 97 | |
| 8729211183 | Electron transport chain occurs in | The inner mitochondrial membrane | 98 | |
| 8729221496 | Electron transport chain | Release of energy stored within reduced hydrogen carriers in order to synthesise ATP | 99 | |
| 8729234126 | Oxidative phosphorylation | Energy to synthesise ATP is derived from the oxidation of hydrogen carriers during the electron transport chain. | 100 | |
| 8729242057 | 3 steps of Oxidative phosphorylation | 1) Generating a Proton Motive Force 2) ATP Synthesis via Chemiosmosis 3) Reduction of Oxygen | 101 | |
| 8729282350 | Generating a Proton Motive Force (Oxidative phosphorylation) | 1) hydrogen carriers are oxidised and release high energy electrons and protons 2) electrons transfer to electron transport chain, made of several transmembrane carrier proteins 3) Electrons moving through the chain lose energy, which is used by the chain to pump protons (H+ ions) from the matrix 4) Accumulation of H+ ions within intermembrane space creates electrochemical gradient | 102 | |
| 8729282351 | ATP Synthesis via Chemiosmosis (Oxidative phosphorylation) | 1) Proton motive force causes H+ to move down their electrochemical gradient and diffuse back into matrix (chemiosmosis, helped by ATP synthase) 2) As H+ ions move through ATP synthase they trigger the molecule rotation of the enzyme, synthesizing ATP | 103 | |
| 8729348174 | Chemiosmosis | Diffusion of protons | 104 | |
| 8729351703 | ATP synthase | Enzyme facilitating chemiosmosis | 105 | |
| 8729282352 | Reduction of Oxygen (Oxidative phosphorylation) | 1) Removal of de-energized electrons, so the chain can continue to function by oxygen 2) Oxygen also binds with free protons in matric to form water, removing matrix protons maintains hydrogen gradient 3) Without oxygen, hydrogen carriers cannot transfer energised electrons to chain and ATP production is halted | 106 | |
| 8729461508 | Three types of aerobic respiration reactions | Decarboxylation, oxidation and phosphorylation | 107 | |
| 8729470566 | Decarboxylation in cellular respiration | Carbon atoms are removed from the organic molecule (glucose) to form carbon dioxide | 108 | |
| 8729491528 | 4 steps of Oxidation in cellular respiration | 1) Electrons and hydrogen ions are removed from glucose and taken up by hydrogen carriers 2) Hydrogen carriers are oxidised at electron transport chain (where energy is used for ATP) 3) Electrons and hydrogen ions are then taken up by oxygen (reduction) to form water 4) 12 hydrogen carriers are produced and six oxygen molecules are required | 109 | |
| 8729491529 | Phosphorylation in cellular respiration | 1) Energy released from glucose breakdown is used to phosphorylate ADP to make ATP 2) Net total of four ATP molecules produced directly via substrate level phosphorylation 3) Remaining ATP is produced indirectly via electron transport chain | 110 | |
| 8729556667 | Aerobic respiration typically produces net total of ... ATP per molecule of glucose consumed | 2 + 2 + 4 + 6 + 18 + 4 = 36 | 111 | |
| 8729631740 | Powerhouse of the cell | Mitochondrion | 112 | |
| 8729647293 | 5 ways in which the structure of the mitochondrion is adapted to the function it performs | - Outer membrane: Transport proteins enabling shuffling of pyruvate from cytosol - Inner membrane: Electron transport chain and ATP synthase for oxidative phosphorylation - Cristae: Folds which increase the SA:Vol ratio - Intermembrane space: Maximizes hydrogen gradient upon proton accumulation - Matrix: Central cavity containing appropriate enzymes and suitable pH for Krebs cycle | 113 | |
| 8729699982 | Electron tomography | Technique by which the 3-dimensional internal structure of a sample can be modelled | 114 | |
| 8729703758 | 3 steps of Electron tomography | 0) (if biological material: dehydrating or freezing) 1) Repeat imaging of samples using electron microscope 2) After image, sample is tilted to different angle 3) computational reconstruction of 3D representation (tomogram) | 115 | |
| 8729757042 | 3 observations of mitochondria made using electron tomography | 1) Cristae are continuous with the internal mitochondrial membrane 2) The intermembrane space is of a consistent width throughout the entire mitochondrion 3) The relative shape, position and volume of the cristae can change in active mitochondria | 116 | |
| 8710772709 | OILRIG | Oxidation Is Losing electrons (gaining energy) Reduction Is Gaining electrons (losing energy) | 117 | |
| 8710780561 | Gaining energy | Breaking bonds between between ADP and creating ATP with breakdown of glucose | 118 | |
| 8711086699 | 5 steps of respiration | 1) Glycolysis 2) Link reaction 3) Kreb's cycle 4) Electron transport chain 5) Chemiosmosis | 119 | |
| 8711160708 | Mitochondria drawing |  | 120 | |
| 8723702202 | Glycolysis | Splitting of glucose into pyruvate | 121 | |
| 8723706857 | Lysis | Splitting | 122 | |
| 8723708998 | Oxydation in glycolisis | NADH+ --> NADH + H | 123 | |
| 8723710004 | Glycolysis 6 steps | 1) Glucose (Hexose, 6C) 2) Phosphorylation 3) Lysis 4) Oxidation 5) ATP formation 6) Pyruvate (2x) | 124 | |
| 8723722430 | The anaerobic cell respiration | Glycolysis | 125 | |
| 8723724114 | ATP yield of Glycolysis | - 2 ATP + 4 ATP = 2ATTP | 126 | |
| 8723768070 | Glycolysis occurs in ... | Cytoplasm | 127 | |
| 8723771571 | Glycolysis summary | 1) Hexose sugar is phosphorylated using ATP 2) Hexose phosphate is split in 2 triose phosphates 3) Oxidation removes hydrogen 4) Hydrogen is used to reduce NAD to NADH 5) 4 ATP produced resulting in net gain of 2 ATP 6) 2 pyruvate molecules are produced at end | 128 | |
| 8723782703 | Oxidate Decarboxylation diagram | 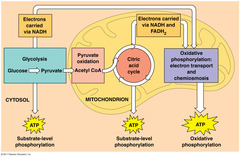 | 129 | |
| 8723789570 | Oxidate Decarboxylation Net Yield | 2 Acetyl CoA per glucose molecule | 130 | |
| 8723792542 | Oxidate Decarboxylation occurs in ... | Mitochondria | 131 | |
| 8723795614 | Oxidate Decarboxylation summary | 1) Pyruvate enters mitochondria matrix 2) Enzymes remove 1xCO2 and H from pyruvate 3) Hydrogen is accepted by NAD to form NADH 4) Removal of hydrogen (oxidation) 5) Removal of carbon dioxide is decarbodylation 6) Link reaction is therefore oxidative decarboxylation 7) Product is acetyl group, reacting w. coenzyme A 8) Acetyl CoA enters Krebs cycle | 132 | |
| 8766353883 | Photosynthesis | Process by which cells synthesise organic molecules (e.g. glucose) from inorganic molecules (CO2 and H2O) in the presence of sunlight | 133 | |
| 8766366410 | Two steps of photosynthesis | 1) Light dependent reactions 2) Light independent reactions | 134 | |
| 8766379834 | Light dependent reactions (3 steps) | 1) Chlorophyll absorbs light and releases energised electrons that are used to produce ATP 2) Carrier molecules (NADP+) gets electrons 3) Photolysis: Water replaces lost electrons in chlorophyll | 135 | |
| 8766413580 | The light dependent reactions occur in the | Thylakoids | 136 | |
| 8766569411 | The light independent reactions occur in the | Stroma | 137 | |
| 8766415647 | Thylakoids | Intermembrane space of membranous discs of chlorophyl | 138 | |
| 8766383007 | Light independent reactions (2 steps) | 1) Transfer of ATP and hydrogen / electrons (Carried by NADPH) to stroma 2) CO2 combines w. H/electrons to form complex organic compounds (Via ATP) | 139 | |
| 8766526098 | Stroma | Fluid-filled interior of the chloroplast | 140 | |
| 8766406484 | Photolysis | Splitting of water into oxygen and hydrogen | 141 | |
| 8766846981 | 3 steps of light dependent reactions to convert light energy into chemical energy (ATP and NADPH) | 1) Excitation of photosystems by light energy 2) ATP Production via an electron transport chain 3) Reduction of NADP+ and the photolysis of water | 142 | |
| 8766865755 | Step 1 of light dependent reaction: Excitation of Photosystems by Light Energy | 2) When a photosystem absorbs light energy, delocalised electrons in pigments get energised 3) These excited electrons are transferred to carrier molecules within the thylakoid membrane | 143 | |
| 8766886895 | Photosystems | Groups of photosynthetic pigments (including chlorophyll) embedded within the thylakoid membrane | 144 | |
| 8766897151 | Photosystems are classed according to | Their maximal absorption wavelengths | 145 | |
| 8766953638 | What generates excited electrons | Absorption of light by photosystems | 146 | |
| 8766962129 | Location of Transfer of excited electrons | Between carriers in thylakoid membranes | 147 | |
| 8767000545 | Step 2 of light dependent reaction: Production of ATP via an Electron Transport Chain | 1) Excited electrons from Photosystem II are transferred to an electron transport chain within the thylakoid membrane 2) As electrons are passed through the chain they lose energy, which is used to translocate H+ ions into the thylakoid 3) Build up of protons in thylakoid creates electrochemical gradient, or proton motive force 4) H+ ions return to stroma via transmembrane enzyme ATP synthase 5) ATP synthase uses passage of H+ ions to catalyse synthesis of ATP (from ADP + Pi) 6) The newly de-energised electrons from Photosystem II are taken up by Photosystem I | 148 | |
| 8767041927 | Photophosphorylation | ATP production by the light dependent reactions | 149 | |
| 8767060893 | Is used to contribute to generate a proton gradient in the light dependent reaction | Excited electrons from Photosystem II | 150 | |
| 8767065820 | Generates ATP using the proton gradient | ATP synthase in thylakoids | 151 | |
| 8767079327 | Step 3 of the light dependent reaction: Reduction of NADP+ and the Photolysis of Water | 1) Excited electrons from Photosystem I may be transferred to a carrier molecule and used to reduce NADP+ --> This forms NADPH 2) The electrons lost from Photosystem I are replaced by de-energised electrons from Photosystem II 3) Electrons lost from Photosystem II are replaced by electrons released from water via photolysis 4) Water is split by light energy into H+ ions and oxygen | 152 | |
| 8767120920 | Two types of Photophosphorylation | Cyclic and non-cyclic processes | 153 | |
| 8767136840 | Cyclic photophosphorylation | Use of only one photosystem (PS I) and does not involve the reduction of NADP+ | 154 | |
| 8767161793 | 3 steps of Cyclic photophosphorylation | 1) Light is absorbed by Photosystem I --> excited electron may enter into electron transport chain to produce ATP 2) De-energized electron returns to photosystem, restoring its electron supply | 155 | |
| 8767181325 | Why is water not needed in the cyclic photophosphorylation and why is NADP+ not reduced | Because the electron returns to the photosystem | 156 | |
| 8767192723 | 3Cyclic photophosphorylation | Photophosphorylation that involves two photosystems (PS I and PS II) and does involve the reduction of NADP+ | 157 | |
| 8767203945 | 3 steps of non-Cyclic photophosphorylation | 1) Light is absorbed by Photosystem II and the excited electrons enter into an electron transport chain to produce ATP 2) Meanwhile, photoactivation of Photosystem I results in the release of electrons which reduce NADP+ (forms NADPH) 3) The photolysis of water releases electrons which replace those lost by Photosystem II (PS I electrons replaced by PS II | 158 | |
| 8767219431 | Cyclic vs Non-Cyclic Photophosphorylation |  | 159 | |
| 8910080250 | The light independent reactions occur in | Stroma (fluid-filled space of chloroplast) | 160 | |
| 8910087369 | Collective name of the light independent reaction | The calvin cycle | 161 | |
| 8910091559 | Three main steps of the calvin cycle/light independent reactions | 1) Carbon fixation 2) Reduction of GP 3) Regeneration of RUBP | 162 | |
| 8910110066 | Carbon fixation | "Carboxylation of ribulose bisphosphate" | 163 | |
| 8910113024 | "Reduction of glycerate-3-phosphate" | 164 | ||
| 8910116969 | Regeneration of ribulose bisphosphate | 165 |

