| 9928771191 | Covalent Bonds | occurs when valence electrons are shared by two atoms | | 0 |
| 9928771192 | Nonpolar covalent bonds | occurs when the electrons being shared are shared equally between the two atoms | | 1 |
| 9928771193 | Polar covalent bonds | one atom has greater electronegativity than the other, resulting in an unequal sharing of the electrons (In h20 - O is slightly negative and H is slightly positive) | | 2 |
| 9928771194 | Ionic Bonds | two atoms attract valence electrons so unequally that the more electronegative atom steals the electron away from the less electronegative atom | | 3 |
| 9928771195 | Ion | resulting charged atom or molecule from an ionic bond | | 4 |
| 9928771196 | Hydrogen bond | weak bonds that for between partial positively charged H atom of one molecule and the strongly electronegatively charged O or N atom of another molecule | | 5 |
| 9928771197 | Van der Waals Interactions | very weak connections that are the result of the asymmetrical distribution of electrons within a molecule. they contribute to the 3D shape of molecules | | 6 |
| 9928771198 | Properties of Water | - One O- and two H+ atoms
- Water molecules are polar
- Hydrogen bonds form between H20 molecules
- maximum of 4 H bonds at a time per water molecule | | 7 |
| 9928771199 | Cohesion | the linking of molecules (ex: bugs can walk on water due to this property) | | 8 |
| 9928771200 | Adhesion | clinging of one substance to another (ex: water droplets adhering to glass windshield) | | 9 |
| 9928771201 | Specific Heat | the amount of heat required to change the temperature of a substance 1 degree celsius. (ex: high specific heat makes the temp. of our oceans stable and able to support plant/animal life) | | 10 |
| 9928771202 | Hydrophilic | water-soluble sustances (ex: sugars, ioinic compounds, some proteins, etc) | | 11 |
| 9928771203 | Hydrophobic | nonpolar substances (ex: oil) that do not dissolve in water | | 12 |
| 9928771204 | pH scale | acidic/basic conditions affect living organisims |  | 13 |
| 9928771205 | Major elements of life | C, H, O, N, S, and P | | 14 |
| 9928771206 | Isomers | molecules that have the same molecular formula but differ in their arrangement of these atoms. (ex: glucose and fructose have same omolecular formula but different roles) | | 15 |
| 9928771207 | Hydroxyl (-OH) Functional Group | - ex: ethanol, methanol
- helps dissolve sugars |  | 16 |
| 9928771208 | Carboxyl (-COOH) Functional Group | - C double bonded to O and has bond to OH
- ex: fatty acids, sugars |  | 17 |
| 9928771209 | Carbonyl (| - C double bonded to O
- ex: ketones and aldehydes such as sugars |  | 18 | |
| 9928771210 | Amino (-NH2) Functional Group | - ex: amino acids |  | 19 |
| 9928771211 | Phosphate (PO3) Functional Group | - ex: organic phosphate, including ATP, DNA, and phospholipids |  | 20 |
| 9928771212 | Sulfhydryl (-SH) Functional Group | - ex: some amino acids, forms disulfide bridges in proteins |  | 21 |
| 9928771213 | Methyl (-CH3) | - ex: addition of a methyl group affects the expression of genes |  | 22 |
| 9928771214 | Polymers | long chain molecules made of repeating subunits called monomers (ex: starch is a polymer composed of glucose monomers) | | 23 |
| 9928771215 | Dehydration reactions/synthesis | creat polymers from monomers. Two monomers are joined by removing one molecule of water | | 24 |
| 9928771216 | Hydrolysis | occurs when water is added to split large molecules | | 25 |
| 9928771217 | Carbs | includes simple sugars (glucose, fructose, etc.) and polymers such as starch. | | 26 |
| 9928771218 | Monosaccharides | monomers of carbohydrates (ex: glucose and ribose) |  | 27 |
| 9928771219 | Polysaccharides | polymers of monosaccharides (ex: starch[plants], cellulose, glycogen [animals]) |  | 28 |
| 9928771220 | Lipids | - all hydrophobic. they are not polymers because they are assembled from a variety of components (ex: waxes, oils, fats, steriods) | | 29 |
| 9928771221 | Fats (triglycerides) | - made up of glycerol molecule and three fatty acid molecules | | 30 |
| 9928771222 | Fatty Acids | nonpolar, hydrocarbon chains that are hydrophobic | | 31 |
| 9928771223 | Saturated Fatty Acids | - have no double bond between carbons
- are solid at room temp
- ex: butter, lard |  | 32 |
| 9928771224 | Unsaturated Fatty Acids | - have double bond between carbons that results in kink
- liquid at room temp
- ex: corn oil and olive oil |  | 33 |
| 9928771225 | Phospholipids | - make up cell membrane
- have hydrophilic (polar) head
- two fatty acids tails which are hydrophobic |  | 34 |
| 9928771226 | Steroids | - made up of 4 rings fused together
- ex: cholesterol is a steriod. it is a common component of cell membranes
- estrogen and testosterone are steroid hormones |  | 35 |
| 9928771227 | Proteins | polymers made up of amino acids monomers | | 36 |
| 9928771228 | Amino Acids | - central carbon bonded to carboxyl group (COOH)
- an amino group (NH2)
- an H atom
- and an R group |  | 37 |
| 9928771229 | Peptide Bonds | Link amino acids. Formed by dehydration synthesis between amino and carboxyl groups of adjacent monomers | | 38 |
| 9928771230 | 4 Levels of Protein Structure | - Primary
- Secondary
- Tertiary
- Quaternary | | 39 |
| 9928771231 | Primary Structure | the unique sequence in which amino acids are joined | | 40 |
| 9928771232 | Secondary Structure | either the alpha helix (coiled, slinky shape) or beta pleated sheet (accordion shape) 3D shape. this is the result from H bonds between polypeptide backbone | | 41 |
| 9928771233 | Tertiary Structure | results in complex shape due to interactions between R groups | | 42 |
| 9928771234 | Quaternary Structure | association of two or more polypeptide chains into one large protein | | 43 |
| 9928771235 | Denaturation | when a protein loses its shape and ability to function due to heat, a change in pH, etc. | | 44 |
| 9928771236 | Nucleotides | - Nitrogenous base (A, T, C, G in DNA - A, U, C, G in RNA)
- Pentose (5 carbon) sugar
- Phosphate group | | 45 |
| 9928771237 | Plasma Membrane | boundary for cell - selectively permeable | | 46 |
| 9928771238 | 3 things about eukaryotic cells | 1) membrane-enclosed nucleus contains chromosomes
2) membrane-bound organelle in cytoplasm
3) euk. much bigger than prok. | | 47 |
| 9928771239 | Nuclear envelope | double membrane surrounding nucleus | | 48 |
| 9928771240 | Nucleolus | region in nucleus where rRNA complexes with proteins to form ribosomal subunits | | 49 |
| 9928771241 | Endoplasmic Reticulum | network of membranes and sacs that takes up more than half of membrane structure |  | 50 |
| 9928771242 | Smooth E.R. | -synthesis of lipids
-metabolism of carbs.
-detoxification of drugs and poisons | | 51 |
| 9928771243 | Rough E.R. | -ribosomes on surface
-synthesize proteins
-proteins move from rough E.R. to golgi | | 52 |
| 9928771244 | Golgi Apparatus | (postal system analogy)
proteins are modified, stored, shipped |  | 53 |
| 9928771245 | Lysosomes | sacs of enzymes that can digest molecules |  | 54 |
| 9928771246 | Vacuoles | store food/water for protists |  | 55 |
| 9928771247 | Central Vacuole | in plant cell, store water | | 56 |
| 9928771248 | Mitochondria | -site of cellular respiration
-found in both plant and animal cells |  | 57 |
| 9928771249 | Chloroplasts | the site of photosynthesis in plants |  | 58 |
| 9928771250 | Cytoskeleton | network of protien fibers through cytoplasm for support, mobility, and regulation | | 59 |
| 9928771251 | Centrosomes | region near nucleus where microtubules grow |  | 60 |
| 9928771252 | Centrioles | located within centrosomes | | 61 |
| 9928771253 | Flagella | help to propel through water (ex: sperm) |  | 62 |
| 9928771254 | Isotonic Solution | no net movement of water | | 63 |
| 9928771255 | Hypertonic Solution | cell will lose water to its surroundings, may shrivel and die (more solute in water than in cell) ex: cell and ocean water | | 64 |
| 9928771256 | Hypotonic Solution | water enters cell faster than it leaves, cell may swell and burst (ex: cell and distilled water) | | 65 |
| 9928771257 | Facilitated Diffusion | the process by which ions and hydrophilic solutions diffuse across the cell membrane with the help of proteins (dont need ATP) |  | 66 |
| 9928771258 | Active Diffusion | substances move against concentration gradient (from less concentrated to more concentrated) this required atp (ex: NA-K pump) |  | 67 |
| 9928771259 | Endocytosis | cell forms new vesicles from plasma membrane to take in molecules (ex: white blood cells engulfing foreign particles) |  | 68 |
| 9928771260 | Exocytosis | Vesicles from interior fuse with cell membrane to expel contents inside cell |  | 69 |
| 9928771261 | 3 stages of cell communication | 1) Reception
2) Transduction
3) Responce | | 70 |
| 9928771262 | Apoptosis | controlled cell suicide to protect neighboring cells from damage that could occur | | 71 |
| 9928771263 | Somatic Cell | any human cell that is not a sex cell (46 chromosomes) | | 72 |
| 9928771264 | Gamete | sex cells like sperm and egg (haploid - 23 chromosomes) | | 73 |
| 9928771265 | Interphase (mitosis) | 1) G1 Phase - growth, checkpoint (if bad --> G0)
2) S Phase - duplicated chromosomes
3) G2 Phase - growth, checkpoint | | 74 |
| 9928771266 | Mitosis Prophase | 1) chromatin becomes chromatids
2) nucleoli disappears
3) mitotic spindle begins to form | | 75 |
| 9928771267 | Mitosis Prometaphase | 1) nuclear envelope fragments
2) chromatids held to each other by centromere | | 76 |
| 9928771268 | Mitosis Metaphase | 1) chroms. move to metaphase plate at equator
2) centrioles are at opposite poles | | 77 |
| 9928771269 | Mitosis Anaphase | 1) chromosomes separate
2) cell elongates
3) by the end, opposite ends of cell contain complete sets of chroms. | | 78 |
| 9928771270 | Mitosis Telophase | 1) nuclear envelope reforms
2) cytokinesis begins (in animals, cleavage furrow forms - in plants, cell plate forms) | | 79 |
| 9928771271 | Binary Fission | how prokaryotes (bacteria) replicate their genome rather than mitosis | | 80 |
| 9928771272 | Major cell checkpoints | g1, g2, m phase checkpoints | | 81 |
| 9928771273 | Kinases | protein enzymes that control the cell cycle but only active when theyre connected to cyclin proteins (cdks) | | 82 |
| 9928771274 | Transformation | process that converts a normal cell to a cancer cell | | 83 |
| 9928771275 | Metastasis | occurs when cells separate from tumor and enter blood/lymph vessels and travel around body | | 84 |
| 9928771276 | Catabolic pathway | release of energy by the breakdown of compounds (ex: occurs when digestive enzymes break down food and release energy) | | 85 |
| 9928771277 | Anabolic pathway | consume energy to build molecules (ex: occurs when amino acids are linked to form muscle protein during exercise) | | 86 |
| 9928771278 | energy | the capacity to do work | | 87 |
| 9928771279 | thermodynamics | study of energy transformations that occur in matter | | 88 |
| 9928771280 | △G | symbol for change in free energy | | 89 |
| 9928771281 | exergonic reaction | energy is released | | 90 |
| 9928771282 | endergonic reaction | requires energy | | 91 |
| 9928771283 | energy coupling | the use of an exergonic process to drive an endergonic one | | 92 |
| 9928771284 | ATP (adenosine triphosphate) | nitrogenous base (adenine)
ribose
chain of three phosphate groups | | 93 |
| 9928771285 | ADP (adenosine diphosphate) | when atp transfers one phosphate group through hydrolysis it becomes adp | | 94 |
| 9928771286 | catalysts | substances that can change the rate of a reaction without being altered | | 95 |
| 9928771287 | active site | the part of the enzyme that binds to the substrate | | 96 |
| 9928771288 | fermentation | partial degradation of sugars that occur without use of O2 | | 97 |
| 9928771289 | aerobic respiration | most efficient catabolic pathway - O2 is consumed as a reactant | | 98 |
| 9928771290 | Cellular Respiration EQ | C6H12O6 + 6O2 ⥤ 6CO2 + 6H20 + energy | | 99 |
| 9928771291 | oxidation | when a reactant loses one or more electrons and energy | | 100 |
| 9928771292 | reduction | when a reactant gains one or more electrons and energy | | 101 |
| 9928771293 | Glycolysis | - occurs in cytosol
- glucose (6C) is broken down into two pyruvate molecules (3C)
- produces: 2 ATP and 2 NADH | 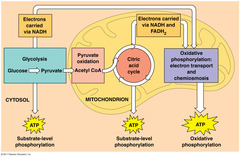 | 102 |
| 9928771294 | Cellular Respiration 4 stages | 1) Glycolysis
2) Pyruvate Oxidation
3) Krebs Cycle
4) Electron Transport Chain | | 103 |
| 9928771295 | Pyruvate Oxidation | - pyruvate oxidized to acetyl CoA
- this releases CO2
- this is a branching point - if cell doesn't have O2 then fermentation occurs, if cell does then krebs cycle occurs | | 104 |
| 9928771296 | Krebs Cycle (aka Citric Acid Cycle) | - in mitochondrial matrix
- releases CO2 as waste
- glucose is broken down
- step-wise catabolism of 6C citrate molecules
- cycle happens twice for each glucose molecule
- produces: 2 ATP, 6 NADH, 4 CO2, 2 FADH2 |  | 105 |
| 9928771297 | Chemiosmosis | - energy-coupling mechanism that stores energy as an H+ gradient to drive cellular work
- electron transport chain and chemiosmosis make up oxidative phosphorylation | | 106 |
| 9928771298 | Anaerobic Respiration | used by prokaryotes to generate ATP without O2 using electron transport chain | | 107 |
| 9928771299 | Fermentation | expansion of glycolysis where atp is generated by substrate-level phosphorylation | | 108 |
| 9928771300 | Alcoholic Fermentation | pyruvate converted to ethanol, releasing CO2 and oxidizing NADH in the process to make more NAD+ | | 109 |
| 9928771301 | Lactic Acid Fermentation | pyruvate is reduced by NADH and lactate is formed as a waste product | | 110 |
| 9928771302 | Stroma/Thylakoid/Chlorophyll/Granum | - Chloroplast is overall structure
- Strome is space inside
- Thylakoids are the connected sacs
- Granum is a stack of thylakoids
- Chlorophyll is located in the thylakoid membranes |  | 111 |
| 9928771303 | Stomata | tiny pores in leaf for CO2 to leave and O2 and H20 leave | | 112 |
| 9928771304 | Photosynthesis | 6CO2 + 6H20 + light ⥤ C6H12O6 + 6O2 | | 113 |
| 9928771305 | Photosynthesis 2 Stages | 1) light reactions
2) Calvin cycle | | 114 |
| 9928771306 | Light Reactions | - light-dependent reaction
- convert solar energy to chemical energy
- ATP and NADPH
- electron transport chain
- protein H+ gradient across inner membrane (PS I and PS II)
- in thylakoid
- H20 goes in and 02 comes out | | 115 |
| 9928771307 | Calvin Cycle | - light-independent reactions
- in stroma
- carbon enters as CO2 and leaves as sugar |  | 116 |
| 9928771308 | Steps of Calvin Cycle | 1) CO2 attaches to RuBP (5C molecule - enzyme rubisco catalyzes reaction)
2) this 6C unstable molecule breaks into PGA
3) ATP phosphorylates PGA, PGA is reduced by NADPH and forms G3P
4) 2 molcules of G3P bond to become one molecule of glucose
5) remaining G3P molecules are phosphorylated by ATP and are regenerated into RuBP | | 117 |
| 9928771309 | Leading strand from ___ to ___ | 5' - 3' | | 118 |
| 9928771310 | Lagging strand from ___ to ___ | 3' - 5' | | 119 |
| 9928771311 | DNA Ligase | seals Okazaki fragments | | 120 |
| 9928771312 | Transcription | the synthesis of RNA using DNA as a template | | 121 |
| 9928771313 | Messenger RNA (mRNA) | carries genetic material to ribosome | | 122 |
| 9928771314 | Translation | the production of a polypeptide chain using the mRNA transcripts (occurs in ribosome) | | 123 |
| 9928771315 | RNA polymerase | enzyme that separates the 2 DNA strands and connects the RNA nucleotides along the the DNA template strand | | 124 |
| 9928771316 | Codons | mRNA base triplets |  | 125 |
| 9928771317 | Promoter | the DNA sequence where the RNA polymerase attaches | | 126 |
| 9928771318 | Terminator | the DNA sequence that signals the end of transcription | | 127 |
| 9928771319 | Stages of Transcription | 1) Initiation
2) Elongation
3) Termination | | 128 |
| 9928771320 | 5' cap and poly-A-tail | help mRNA leave nucleus, protects mRNA from degredation, facilitate attachments of mRNA to ribosome | | 129 |
| 9928771321 | Introns | Part of mRNA that are spliced out through RNA splicing | | 130 |
| 9928771322 | Exons | the sections that remain after RNA splicing | | 131 |
| 9928771323 | Anticodon | codon on tRNA that binds with mRNA | | 132 |
| 9928771324 | tRNA | transfers amino acids from a pool of amino acids in cytoplasm to ribosome | | 133 |
| 9928771325 | point mutation | alteration of just one base pair | | 134 |
| 9928771326 | missense mutation | amino acid the codon codes for is still the same | | 135 |
| 9928771327 | nonsense mutation | amino acid changes to code for a stop codon | | 136 |
| 9928771328 | 3 parts of operon | 1) operator to control access of RNA
2) promoter where RNA polymerase attaches
3) genes of operon | | 137 |
| 9928771329 | restriction enzymes | used to cut strands of DNA at specific locations (restriction sites) | | 138 |
| 9928771330 | Meiosis Interphase | chromosomes duplicate, replicating their DNA. centrosome also divides | | 139 |
| 9928771331 | Meiosis I Prophase | -chromosomes condense, resulting in 2 sister chromatids attached at their centromeres
-crossing over (synapsis) occurs (now have tetrads and chismatas)
-spindle poles move away from each other, nuclear envelope goes away, spindle microtubules attach to kinetochores on chroms. |  | 140 |
| 9928771332 | Meiosis I Metaphase | -lined up at metaphase plate |  | 141 |
| 9928771333 | Meiosis I Anaphase | -spindle apparatus helps to move chroms to opposite ends of cell
-sister chroms stay connected, homologous chroms separate |  | 142 |
| 9928771334 | Meiosis I Telophase and Cytokinesis | -each pole has haploid set of chroms, each chrom consisting of 2 sister chromatids
-cleavage furrow forms in animals, cell plates in plant cells
-daughter cells are now haploid |  | 143 |
| 9928771335 | Meiosis II Prophase | -spindle apparatus forms
-sister chroms move toward metaphase plate |  | 144 |
| 9928771336 | Meiosis II Metaphase | -haploid number of chroms. is now arrayed on metaphase plate
-kinetochores of each sister chromatid are attached to microtubules from opposite poles |  | 145 |
| 9928771337 | Meiosis II Anaphase | -centromeres of sister chromatids separate and individual chroms move to opposite ends |  | 146 |
| 9928771338 | Meiosis II Telophase and Cytokinesis | -nuclei reappear
-each 4 daughter cells has haploid number of chroms | | 147 |
| 9928771339 | Meiosis vs Mitosis | -synapsis doesn't occur during mitosis
-tetrads at metaphase plate rather than individual chroms like in mitosis
-in miosis duplicated homo. chroms separate but sister chroms. stay attached. in mitosis, the chromatids separate | | 148 |
| 9928771340 | Complete dominance | when the heterozygote and homosygote for the dominant allele are the same
ex: Yy and YY are both equally yellow | | 149 |
| 9928771341 | Codominance | occurs when 2 alleles are dominant and affect the phenotype in two different, equal ways
ex: blood types - A and B are dominant to O but A and B are codominant to each other | | 150 |
| 9928771342 | Incomplete Dominance | where Fi hybrids have an appearance in between that of 2 parents
ex: red flower and white flower breed and make pink | | 151 |
| 9928771343 | Polyploidy | having more that two complete sets of chroms. (rare in animals, frequent in plants) | | 152 |
| 9928771344 | Chromosome Deletion | when chrom. fragment is lost (missing genes) | | 153 |
| 9928771345 | Chromosome Duplicaiton | when chrom. segment is repeated | | 154 |
| 9928771346 | Chromosome Inversion | when chrom. fragment breaks off and reattaches backward |  | 155 |
| 9928771347 | Chromosome Translocation | when chrom. fragments breaks off and reattaches on a nonhomologous chrom. | | 156 |
| 9928771348 | p + q = 1 | p= frequency of dominant alleles in pop.
q= frequency of recessive alleles in pop. | | 157 |
| 9928771349 | p^2+2pq+q^2=1 | p^2= frequency of homo. dominant individuals
q^2= frequency of homo. recessive individuals
2pq= frequency of heterozygous individuals | | 158 |
| 9928771350 | antibody | a blood protein produced in response to antigen | | 159 |
| 9928771351 | antigen | a toxin or other foreign substance that induces an immune response in the body | | 160 |
| 9928771352 | b-cell | a lymphocyte responsible for producing antibodies | | 161 |
| 9928771353 | cell-mediated immunity | an immune response that does not involve antibodies, but involves the activation of phagocytes, cytotoxic T-lymphocytes, and cytokines in response to an antigen | | 162 |
| 9928771354 | cell communication | the process by which a cell detects and responds to signals in its environment | | 163 |
| 9928771355 | cyclic AMP (cAMP) | lays a major role in controlling many enzyme-catalyzed processes |  | 164 |
| 9928771356 | cytotoxic t-cell (killer t-cells) | is a T lymphocyte (a type of white blood cell) that kills cancer cells, cells that are infected (particularly with viruses), or cells that damaged in other ways | | 165 |
| 9928771357 | g-protein linked receptor | detect molecules outside the cell and activate internal signal |  | 166 |
| 9928771358 | helper t-cell | a T cell that influences or controls the differentiation or activity of other cells of the immune system | | 167 |
| 9928771359 | hormone | a regulatory substance produced in an organism (transported in tissue fluids such as blood) to stimulate specific cells into action | | 168 |
| 9928771360 | humoral immunity | aspect of immunity that is mediated by macromolecules found in extracellular fluids (body fluids) such as secreted antibodies, complement proteins, and certain antimicrobial peptides | | 169 |
| 9928771361 | inducer | a molecule that regulates gene expression. An inducer can bind to protein repressors or activators. Inducers function by disabling repressors. The gene is expressed because an inducer binds to the repressor |  | 170 |
| 9928771362 | lytic cycle | one of the two cycles of viral reproduction, the other being the lysogenic cycle. The lytic cycle results in the destruction of the infected cell and its membrane. | | 171 |
| 9928771363 | operon | a segment of DNA to which a transcription factor binds to regulate gene expression by repressing it. Repressors bind to operators to prevent transcription | | 172 |
| 9928771364 | phagocyte | cells that protect the body by ingesting (phagocytosing) harmful foreign invaders | | 173 |
| 9928771365 | phosphorylation cascade | a sequence of events where one enzyme phosphorylates another, causing a chain reaction leading to the phosphorylation of thousands of proteins |  | 174 |
| 9928771366 | protein kinase | a kinase enzyme that modifies other proteins by chemically adding phosphate groups to them (phosphorylation) | | 175 |
| 9928771367 | receptor | A molecular structure or site on the surface or interior of a cell that binds with substances such as hormones, antigens, drugs, or neurotransmitters |  | 176 |
| 9928771368 | repressor | a DNA- or RNA-binding protein that inhibits the expression of one or more genes by binding to the operator |  | 177 |
| 9928771369 | signal transduction | A set of chemical reactions in a cell that occurs when a molecule, such as a hormone, attaches to a receptor on the cell membrane |  | 178 |
| 9928771370 | transcription factor | is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence | | 179 |
| 9928771371 | virus | an infective agent that typically consists of a nucleic acid molecule in a protein coat and is able to live only within the living cells of a host | | 180 |
| 9928771372 | function of enzymes | catalysts for biochemical reactions. They speed up reactions by providing an alternative reaction pathway of lower activation energy. | | 181 |
| 9928771373 | Lyell | idea that shaping of earth took place over long period of time (earth must be very old) | | 182 |
| 9928771374 | Lamarck | characteristics acquired during organisms lifetime is passed down to offspring (ex: giraffe necks) and body parts that are used grow strong those that don't deteriorate | | 183 |
| 9928771375 | homologous structures | anatomical signs of evolution (ex: arms in bats, whales, humans) | | 184 |
| 9928771376 | vestigial organs | organs that have no importance to body (ex: pelvic bone in snake, whale) | | 185 |
| 9928771377 | convergent evolution | explains why distantly related species resemble one another. evolved from similar environments but not similar ancestors (ex: dolphins and fish) | | 186 |
| 9928771378 | endemic species | found at certain geographic locations and nowhere else | | 187 |
| 9928771379 | gene pool | all of the alleles at all loci in all members of a population | | 188 |
| 9928771380 | 5 conditions for H-W Equilibrium | 1. no mutations
2. random mating
3. no natural selection
4. population must be large
5. no gene flow (emigration, immigration...) | | 189 |
| 9928771381 | founder effect (genetic drift) | a few individuals become isolated from a larger population and establish new one | | 190 |
| 9928771382 | bottleneck effect (genetic drift) | sudden change in environment (natural disaster) that reduces pop. | | 191 |
| 9928771383 | directional selection | extreme phenotype favored over others |  | 192 |
| 9928771384 | disruptive selection | 2 extreme values favored over middle |  | 193 |
| 9928771385 | stabilizing selection | favors intermediate values |  | 194 |
| 9928771386 | speciation | process by which new species arise | | 195 |
| 9928771387 | microevolution | change in genetic makeup of a pop from gen. to gen. | | 196 |
| 9928771388 | macroevoltuon | broad pattern of evolutionary change above species levels | | 197 |
| 9928771389 | biological species concept | defines a species as a group of populations whose members have the potential to produce offspring, but cannot produce offspring with other species | | 198 |
| 9928771390 | Habitat isolation (prezygotic) | two species live in same area but not same habitat (ex: one wrong lives in tree, one on ground) | | 199 |
| 9928771391 | Behavioral Isolation (prezygotic) | signals to attract mates that are unique to their species | | 200 |
| 9928771392 | Temporal Isolation (prezygotic) | species breed at different times of day/seasons/years (ex: flowers in spring only vs. fall only) | | 201 |
| 9928771393 | Mechanical Isolation (prezygotic) | species are anatomically incompatible (ex: ant and horse) | | 202 |
| 9928771394 | Gamete Isolation (prezygotic) | gametes are unable to fuse to form a zygote (ex: dog and bunny) | | 203 |
| 9928771395 | Reduces hybrid viability (postzygotic) | when zygote is formed, genetic incompatibility causes development to cease | | 204 |
| 9928771396 | reduced hybrid fertility (postzygotic) | even if offspring is produced, reproduction isolation still occurs if offspring is sterile | | 205 |
| 9928771397 | hybrid breakdown (postzygotic) | two species mate and have viable offsprings but offspring is weak/sterile | | 206 |
| 9928771398 | allopatric speciation | a population is separated and two different species are formed | | 207 |
| 9928771399 | sympatric speciation | small part of pop. forms new species without being geographically isolated (ex: polyploidy plant) | | 208 |
| 9928771400 | adaptive radiation | occurs when many new species arise from one common ancestor | | 209 |
| 9928771401 | gradualism | species descended from common ancestor and gradually change | | 210 |
| 9928771402 | punctuated equilibruim | periods of stasis punctuated by sudden change | | 211 |
| 9928771403 | oligotrophic lakes | deep lakes, nutrient poor and oxygen rich- contain sparse phytoplankton | | 212 |
| 9928771404 | eutrophic lakes | shallow, high nutrition content and low oxygen content - lots of phytoplankton | | 213 |
| 9928771405 | biotic factors | behaviors and interactions of species | | 214 |
| 9928771406 | abiotic factors | environment, climate, non-living factors | | 215 |
| 9928771407 | k-selection | -late reproduction
-few offspring
-invest in raising offspring
-logistic growth | | 216 |
| 9928771408 | r-selection | -early reproduction
-many offspring
-little parental care
-expontential growth | | 217 |
| 9928771409 | Competition (-/-) | when resources are short in supply | | 218 |
| 9928771410 | Predation (+/-) | one species is predator and one is prey | | 219 |
| 9928771411 | cryptic coloration | animal is camoflaged by its coloring | | 220 |
| 9928771412 | aposematic (warning coloration) | poisonous animal is brightly colored to warn other animals | | 221 |
| 9928771413 | batesian mimicry | harmless species mimics harmful species | | 222 |
| 9928771414 | mullerian mimicry | two bad-tasting species mimic each other so predators avoid them both equally | | 223 |
| 9928771415 | Symbiosis | when individuals of two or more species live in direct contact with one another | | 224 |
| 9928771416 | Parasitism (+/-) | parasite derives nutrients from host | | 225 |
| 9928771417 | Mutualism (+/+) | benefits both species (ex: bees and flowers0 | | 226 |
| 9928771418 | Commensalism (+/0) | benefits one species and neither helps nor hurts another (ex: fern growing in shade of another plant) | | 227 |
| 9928771419 | biomass | (sum weight of all members of a pop.) | | 228 |
| 9928771420 | primary production | amount of light energy converted to chemical energy by autotrophs | | 229 |
| 9928771421 | gross primary production | total primary production in ecosystem | | 230 |
| 9928771422 | net primary production | gpp - respiration | | 231 |
| 9928771423 | eutrophic | a lake that is nutrient rich and supports a vast amount of algae | | 232 |
| 9928771424 | nitrification | ammonium (NH4+) is oxidized to nitrite and then nitrate by bacteria | | 233 |
| 9928771425 | dentrification | releases nitrogen to atmosphere by bacteria | | 234 |
| 9928771426 | bioremediation | use of organisms to detoxify polluted ecosystem | | 235 |
| 9928771427 | osmosis | movement of water from high concentration to low concentration through selectively permeable membrane | | 236 |
















































































