| 7020172606 | plasma membrane | separates internal metabolic event; controls mvmnt of materials |  | 0 |
| 7020172607 | saturated fatty acid membrane | packed tight; rigid membrane |  | 1 |
| 7020172608 | unsaturated fatty acid membrane | not packed tight; flexible membrane |  | 2 |
| 7020172609 | selectively permeable | allows: small, unchanged, polar molecules; hydrophobic molecules
blocks: large polar molecules; ions |  | 3 |
| 7020172610 | integral proteins | imbedded in bilipid layer |  | 4 |
| 7020172611 | peripheral proteins | attached to membrane surface |  | 5 |
| 7020172612 | channel proteins | passage for hydrophillic substances |  | 6 |
| 7020172613 | aquaporins | channel proteins; increase rate of H20 passage |  | 7 |
| 7020172614 | ion channels | ions; gated channels |  | 8 |
| 7020172615 | gated channels | open and close in response to stimuli
EX: nerve and muscle cells |  | 9 |
| 7020172616 | carrier proteins | specific molecules bind, changing protein shape |  | 10 |
| 7020172617 | transport proteins | use ATP (active transport)
EX: sodium-potassium pump |  | 11 |
| 7020172618 | recognition proteins | unique identification; glycoproteins
EX: blood types |  | 12 |
| 7020172619 | receptor proteins | provide binding sites; activates specific cell response |  | 13 |
| 7020172620 | adhesion proteins | attaches cells to cells; provides anchors for filaments |  | 14 |
| 7020172621 | cholesterol | stability to animal cells; @ high T=maintain firmness, @ low T=allows flexibility |  | 15 |
| 7020172622 | organelles | bodies within cytoplasm; chemical rxns isolated, able to take place w/o interference; large surface areas to max. space for rxns | 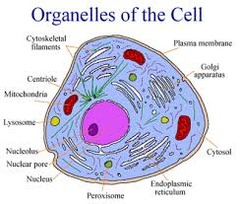 | 16 |
| 7020172623 | nucleus | contains DNA and nucleoli; site of cell division |  | 17 |
| 7020172624 | chromatin | DNA spread through nucleus like web |  | 18 |
| 7020172625 | chromosomes | DNA becomes rod-shaped as cell begins division; includes 2 long DNA molecules and histone proteins |  | 19 |
| 7020172626 | histones | organize long DNA |  | 20 |
| 7020172627 | nucleosomes | coiling of DNA by histones |  | 21 |
| 7020172628 | nucleoli | concentrations of DNA in process of manufacturing ribosomes |  | 22 |
| 7020172629 | nuclear pores | passageway for proteins and RNA |  | 23 |
| 7020172630 | nuclear envelope | 2 bilipid layers; bounded to nucleus, continuous with ER |  | 24 |
| 7020172631 | ribosome | 2 RNA subunits + proteins; free v bound; protein synthesis |  | 25 |
| 7020172632 | free ribosomes | in cytosol; proteins made function within cell
EX: enzymes that catalyze sugar breakdown |  | 26 |
| 7020172633 | bound ribosomes | attached to ER or nuclear envelope; proteins made function within cell membrane or exported from cell |  | 27 |
| 7020172634 | smooth ER | w/o ribosomes; synthesis of lipids and steroids, metabolizes CHO, detoxification |  | 28 |
| 7020172635 | rough ER | w/ ribosomes; synthesis of proteins and glycoproteins, produces new membrane |  | 29 |
| 7020172636 | golgi apparatus | collect, modify, and package proteins, CHO and lipids |  | 30 |
| 7020172637 | lysosomes | (animal cells only) vesicles from Golgi with hydrolytic enzymes; break down material in cytosol for recycling; low pH |  | 31 |
| 7020172638 | peroxisomes | animals: breakdown H202, fatty acids, AAs; plants: modify by-products of photosynthesis |  | 32 |
| 7020172639 | mitochondria | carry out cellular respiration; two membranes allow separation of metabolic processes |  | 33 |
| 7020172640 | chloroplasts | (plant cells only) carry out photosynthesis; two membranes |  | 34 |
| 7020172641 | microtubules | made of protein tubulin; found in spindle apparatus (guides chromosome mvmnt in cell division); support and motility for cell activity |  | 35 |
| 7020172642 | intermediate filaments | support for cell shape |  | 36 |
| 7020172643 | microfilaments | made of protein actin; found in cells that move by shape change, e.g. muscle cells; cell motility |  | 37 |
| 7020172644 | flagella | long, few, snake-like mvmnt; 9+2 microtubule
EX: sperm |  | 38 |
| 7020172645 | cilia | short, many, back-and-forth mvmnt; 9+2 microtubule
EX: line respiratory tract |  | 39 |
| 7020172646 | centrioles | (animal cells only) microtubule organizing centers; create spine apparatus in cell division |  | 40 |
| 7020172647 | transport vesicles | move materials btwn organelles |  | 41 |
| 7020172648 | food vacuoles | receive nutrients; usually merge with lysosomes |  | 42 |
| 7020172649 | contractile vacuoles | collect and pump water in cell |  | 43 |
| 7020172650 | central vacuoles | (plant cells only) contain most of plant cell interior; exert tugor when full for cell rigidity; functions specialized:
1) store starch, nutrients, waste, etc.
2) lysosome function
3) cell growth by absorbing H20
4) renders large SA-to-V ratio |  | 44 |
| 7020172651 | cell walls | (plant cells only) support |  | 45 |
| 7020172652 | extracellular matrix | (animal cells only) mechanical support, helps bind adjacent cells |  | 46 |
| 7020172653 | cell junctions | anchor cells together for cellular exchange |  | 47 |
| 7020172654 | anchoring junctions | (animals) protein attachments between cells
EX: desmosome |  | 48 |
| 7020172655 | tight junctions | (animals) seal that prevents cellular exchange |  | 49 |
| 7020172656 | communication junctions | allow chemical and electrical signal passage |  | 50 |
| 7020172657 | gap junctions | (animals) (communication) narrow tunnels; basically channel protein connecting 2 cells |  | 51 |
| 7020172658 | plasmodesmata | (plants) (communication) narrow tunnels; like gap junctions |  | 52 |
| 7020172659 | prokaryotes | plasma membrane, DNA, ribosomes, cytoplasm, cell wall |  | 53 |
| 7020172660 | hypertonic | solute hypertonic to solution=higher [solutes] |  | 54 |
| 7020172661 | hypotonic | solute hypotonic to solution=lower [solutes] |  | 55 |
| 7020172662 | isotonic | [solute]=[solution] |  | 56 |
| 7020172663 | bulk flow | collective movement in response to pressure |  | 57 |
| 7020172664 | passive transport | [higher] to [lower]; increases w/ increase in [x], temp., smaller particle size |  | 58 |
| 7020172665 | diffusion | random mvmnt leads to net mvmnt from [high] to [low] |  | 59 |
| 7020172666 | osmosis | diffusion of water across selectively permeable mmbrn |  | 60 |
| 7020172667 | turgor pressure | osmosis into cell |  | 61 |
| 7020172668 | plasmolysis | osmosis out of cell |  | 62 |
| 7020172669 | cell lysis | swelling of cell b/c excess turgor pressure |  | 63 |
| 7020172670 | facilitated diffusion | diffusion through channel or carrier proteins |  | 64 |
| 7020172671 | active transport | [lower] to [higher]; requires use of E (usually ATP) |  | 65 |
| 7020172672 | electrochemical gradient | combo. of concentration and electrical voltage gradients of ions |  | 66 |
| 7020172673 | cotransport | protein that allows downhill mvmt to drive another uphill; E for uphill from [gradient] from downhill |  | 67 |
| 7020172674 | vesicular transport | uses vesicles to move substances across plasma mmbrn |  | 68 |
| 7020172675 | exocytosis | fuse w/ membrane, release contents outside cell |  | 69 |
| 7020172676 | endocytosis | capture substance outside cell, fuse w/ membrane, release contents into cell; 3 types: phagocytosis, pinocytosis, receptor-mediated |  | 70 |
| 7020172677 | phagocytosis | cellular eating; undissolved material enters cell; forms phagocytic vesicle |  | 71 |
| 7020172678 | pinocytosis | cellular drinking; dissolved material enters cell; forms liquid vesicle |  | 72 |
| 7020172679 | receptor-mediated endocytosis | specific molec. (ligands) binds to site, resulting in pinocytosis |  | 73 |
| 7020172680 | water potential | movement of water from where there is high potential to low potential; based upon solute and pressure components |  | 74 |
| 7020172681 | solute potential | =-iCRT
i = ionization constant (NaCl = 2, glucose = 1)
C = concentration (M)
R = constant (0.0831 mol-liters/bar K)
T = temperature (K)
more solute = lower overall potential |  | 75 |
| 7020172682 | pressure potential | measurement of pressure, in an open container usually = 0 |  | 76 |














































































































